Curriculum
In 2023, the Master's program underwent a substantial update, resulting in its present format. Detailed module descriptions are now accessible on this page, alongside the revised study regulations and module handbook. These new regulations are applicable to students enrolling from 2023 onwards.
Students who commenced their studies in 2022 or earlier are governed by the previous study regulations and module handbook.
The diagram below offers a clear overview of the updated program structure, organizing course modules according to their respective semesters. The curriculum is thoughtfully designed to include both compulsory and elective courses, providing students with the flexibility to customize their education to align with their personal and professional interests, thereby enhancing their career prospects.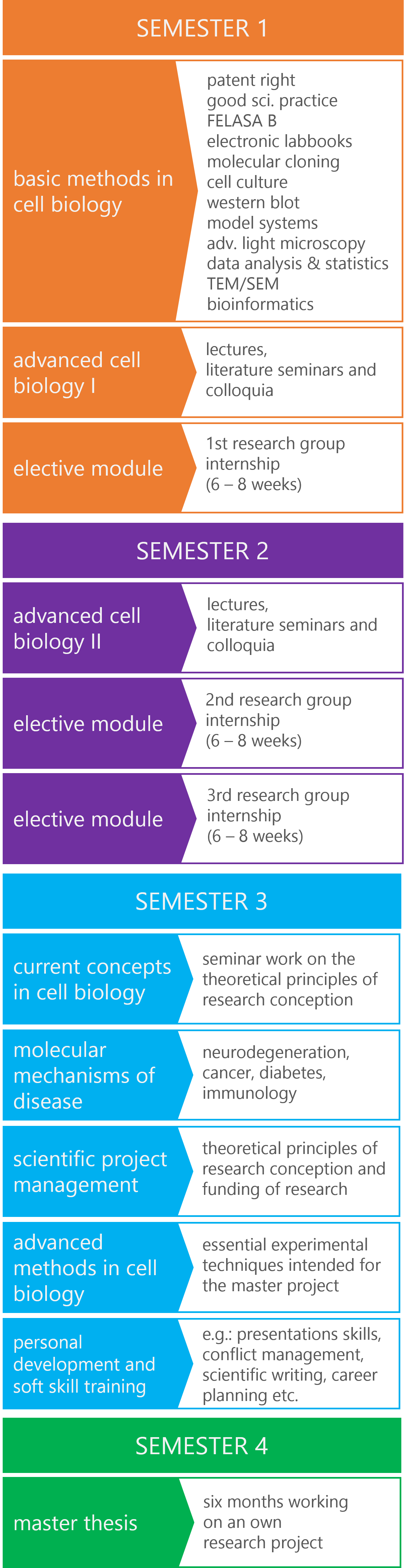
Elective Modules
PD Dr. Bernhard Gaese
Contents
The practical teaches techniques to determine auditory function and dysfunction in rodents. These techniques can be used to determine effects of pharmacological or behavioural treatments of sensory disorders such as tinnitus or hearing loss. The focus is on behavioural techniques suitable to characterize the disorder rather precisely in comparison to normal functions. All steps that are necessary for a project in the field are taught in this practical: study design, animal handling, control of experimental parameters, pharmacological treatment of animals, and data analysis. The behavioural analysis is paralleled by basic electrophysiological measurements necessary to determine the effects of dysfunction and treatments at the physiological level. The students work on their own projects under supervision and present their results in the form of a seminar talk. The main focuses are: measuring and analysing behavioural data, performing efficient physiological experiments to determine auditory function, and statistical evaluation methods. Preparation of a potential publication will be the final part of the project. After completion, the individual projects will be presented and discussed in the form of a seminar talk. In a further seminar talk the students will present an original piece of research from the area of cognition and hearing.Educational objectives / Competences
Familiarity with carrying out well controlled behavioural experiments (animal handling, measuring and analysing behavioural data, statistical analysis). Performing physiological measurements including electrophysiological recording in minimally invasive preparations. Additional aspects are: introduction to software for data handling, signal processing, and graphical display. Deriving scientific questions from the current literature. Knowledge about the usage and limitations of animal models for neurological diseases.
PD Dr. Bernhard Gaese
Contents
The practical covers the whole range of techniques to investigate brain activity underlying the processing of sensory information in the auditory domain. The focus is on electrophysiological single cell techniques in rodents in the awake and anesthetized preparations. Brain activity is acquired and analysed with the goal to understand behavioural responses following auditory stimulation. Cognitive aspects (e.g. context-dependence) are taken into account. The students work on their own projects under supervision and present their results in the form of a seminar talk. The main focuses are measuring and analysing neuronal activity in different configurations of in-vivo recording techniques. The following analysis includes modern techniques of signal processing, efficient handling of larger data sets and statistical evaluation methods. Preparation of a potential publication will be the final part of the project. After completion, the individual projects will be presented and discussed in the form of a seminar talk. In a further seminar talk the students will present an original piece of research from the area of cognition and hearing.Educational objectives / Competences
Familiarity with carrying out physiological experiments (animal handling, surgery, measuring and analyzing electrical activity at the single neuron level. Combining physiology with neuroanatomical and histological staining techniques. Basic introduction to behavioural control. Introduction to software for data handling, signal processing, statistical analysis and graphical display. Understanding cognitive influences on sensory information processing as an important aspect of context-dependent behaviour. Deriving scientific questions from the current literature.
Prof. Dr. Bernd Gruenewald
Contents
The module teaches physiological bases of behaviour control. Students will work on individual projects that are jointly designed. The techniques taught may comprise cell physiology (patch clamp, calcium imaging, extracellular recording), Neuroanatomy, behavioural experiments (behavioural pharmacology, learning and memory). Model organisms are insects, mainly the honey-bee. Conceptional focusses are: function of ion channels and transmitter receptors, neuromodulation, learning behaviour, olfaction, social behaviour of honeybees.The students will give oral presentations of their results and will create a scientific poster summarizing their experiments. In a second seminar talk they learn to critically present physiological and behavioural articles. Presentations, seminars and posters will be given in English and the students receive detailed feedback on their presentation style as well as on the scientific contents. By writing a protocol in a manuscript style the students get acquainted with preparing a manuscript draft for submission to a science journal.
The students will be responsible – under supervision – for the study design, protocolling and analysing the original data. Each step will be developed during the course rather than working after a pre-defined protocol.
Educational objectives / Competences
Planning, conducting and analysing of behavioural physiological experiments; measuring of ionic currents; behavioural observations and quantifications; neuroanatomical methods. Approaching scientific topics; literature work. Preparing of scientific texts, posters and talks.
Prof. Dr. Ernst H.K. Stelzer & Dr. Francesco Pampaloni
Contents
This internship teaches the basic concepts of three-dimensional cell biology and modern three-dimensional microscopy. The observation of live biological specimens under physiologically relevant conditions becomes increasingly important in the Life Sciences. Healthy as well as tumour cell lines, primary cells as well as stem cells, are cultivated and analysed under physiological conditions. These conditions are achieved with organ slices and three-dimensional cellular spheroids by growing them in collagen and many other hydrogels that mimic the extracellular matrix (e.g. Matrigel). Quantitative analyses of living three-dimensional structures requires fast optical sectioning. Confocal microscopy is only useful for relatively thin specimens, because of its slow scanning speed, high photo-bleaching rate and low efficiency in collecting light from thick specimens. Light sheet-based fluorescence microscopy in conjunction with three-dimensional specimen preparation techniques provides a suitable alternative. Students will participate in current research projects of the Stelzer group and are supervised by experienced members. They present their results in an oral presentation and in a written internship report.Educational objectives / Competences
The student learns the basic concepts of classical two-dimensional as well as three-dimensional cell culture. She or he is aware of several applications of three-dimensional cell cultures and knows which cell types are used in the Life Sciences. He or she understands the principles of optics in classical microscopy (characteristics of light, resolution, aperture) as well as photometry (energy, power). The student knows the differences between confocal and light sheet-based fluorescence microscopy and is be able to estimate the limits of classical light microscopy in dense tissues. She or he masters the formation, isolation and staining of spheroids, cysts, organoids and three-dimensional tissue slices. The student has experience in the preparation of the specimens for different microscopes as well as the acquisition and processing of the images and the analysis of the data. At the end of the module the student presents the results in a in written report and a talk.
Prof. Dr. Ernst H.K. Stelzer & Dr. Frederic Strobl
Contents
This internship teaches state-of-the-art three-dimensional fluorescence microscopy and respective non-invasive sample mounting techniques. All steps are shown exemplarily in the investigation of insect embryogenesis. For more than a century, insect research has contributed significantly to genetics and developmental biology. The most prominent model organism is the common fruit fly/vinegar fly Drosophila melanogaster. However, over the last years it became apparent that focusing on a few established model organisms is not sufficient to understand the basic principles of insect development in detail. New insect species (emerging model organisms) are established in many laboratories to gain new insights into neglected or even unknown processes. For example, the red flour beetle Tribolium castaneum is used since its embryogenesis deviates substantially from that of Drosophila in many different aspects. Instead of wide field or confocal fluorescence microscopy, we use light sheet-based fluorescence microscopy, which allows us to image individuals for one week. Moreover, the imaged individual survives the procedure. Students will work on current research projects and are supervised by experienced members of the Stelzer group. They present their results in an oral presentation and a written internship report.Educational objectives / Competences
The student learns the principles of insect model organisms, such as Tribolium, in developmental biology. He or she is aware of current scientific questions in developmental biology and knows how to handle transgenic organisms. He or she understands the principles of optics in classical microscopy (characteristics of light, resolution, numerical aperture) as well as photometry (energy, power). The student knows the differences between confocal and light sheet-based fluorescence microscopy and is be able to estimate the limits of classical light microscopy in dense tissues. He or she understands laboratory cultivation of Tribolium as well as preparation methods for confocal and light sheet-based fluorescence microscopes, in the context of long-term live imaging of Tribolium embryos in toto. The student analyses the data and understands the basics of scientific image processing and the embryonic development of Tribolium.
The interns work under guidance on their own individual project based on the actual research topics of the Stelzer group. At the end of the course, they summarize their results and findings in a protocol and prepare a seminar under the guidance of their advisor.
Prof. Dr. Enrico Schleiff
Contents
The practical course teaches basic techniques and experimental concepts of molecular cell biology and special questions of cellular and molecular aspects of plant physiology.Key features are: protein biochemical methods to study protein translocation and dynamics of chloroplasts, including subcellular fractionation, basics in plant cell culturing and transgenic (genetically modified) plants, in vivo and in situ measurement of protein activity and localization including digital image processing.
The students will learn the handling of genetically modified plants, plant cell cultures and protoplasts e.g. culturing, passaging and transfection for ectopic expression or knockout of genes. The analysis comprises a broad spectrum of molecular biological and cell biological techniques like PCR, cloning, SDS-polyacrylamide gel electrophoresis, western blotting, immunofluorescence, measurement of protein activity and so on.
The students work under supervision on their own scientific project which is leaned on the scientific work of the study group and present their experimental results in form of a seminar lecture. In another lab meeting the students present a recent publication on the field of cellular and molecular plant physiology. By performing a protocol with own scientific results, the students learn to write a scientific paper.
Educational objectives / Competences
Skills taught: Knowledge to isolate plant cell organelles, independent characterization of organelle proteins, sterile working, culturing and transfection of cells, working with the fluorescence microscope and computational evaluation of experimental data and image files, knowledge in the analysis of transgenic plants, independent handling of scientific questions in the context of relevant scientific literature.
Prof. Dr. Ingo Ebersberger
Contents
In this practical we will provide an understanding of basic methods and algorithms for the bioinformatics analysis of large datasets. The students will work on problems circling around the functional characterization and evolution of metabolic pathways and functional protein complexes. We will integrate latest high throughput DNA sequencing data into the analysis whenever possible and appropriate.Emphasis will be put on the compilation of novel sequence data sets for analyses, on data mining for complementation of existing data sets, as well as on the bioinformatics methods for comparison and annotation of sequence data. The theoretical foundation of the analyses will be formed by self-reliant literature research in combination with the presentation of a publication from the area of applied bioinformatics. Towards the end of the internship the students will exercise the correct way of presenting scientific results by summarizing their achievements in an oral presentation as well as in written form in a report.
Educational objectives / Competences
Independent conduct of functional annotation of sequences, of bioinformatics annotation transfer and of prediction of functionally equivalent proteins under consideration of evolutionary relation-ships; Ability for management and bioinformatics analysis of large sequence sets; Mining of public databases; Knowledge of relational database systems; Generation and interpretation of phylogenetic profiles; Introduction into independent scientific research on the background of relevant literature.
PD Dr. Zoe Waibler
Contents
In the practical course the students work on current projects of the working group. Main topics are immunological experiments in the murine system and in primary murine cells as well as primary human cell cultures.In vitro experiments with murine organs and the isolation of murine primary cells will be learned and performed.
The analysis comprises a broad spectrum of immunological and cell culture techniques like: FACS, ELISA, Plaque-Assay, viral Infections, (q)RT-PCR and the culturing of primary human cells and the isolation of different cell types from blood donors as well as the separation of cells with MACS and Cell-Sorter.
Educational objectives / Competences
The students will learn to plan and to perform complex immunological experiments.
The results of the practical course are presented by every student on the form of a written protocol and a talk at the end of the course. The students also take part on the weekly lab meetings where they learn about the ongoing research of all the members of the group. In a Journal Club every student learns to presents a recent publication on the field of immunology and in context of their own projects.
Prof. Dr. Didier Stainier
Contents
This practical course offers basic theoretical and experimental knowledge in the area of developmental genetics. Principal areas of research are the development, function and homeostasis of vertebrate organ systems including the cardiovascular system, lung and pancreas. The students take part on ongoing experiments in the laboratory to elucidate the cellular and molecular mechanisms underlying these processes. Their work includes: basic zebrafish or mouse genetics techniques and the handling of a zebrafish or mouse colony, live imaging of zebrafish embryos and larvae, processing of embryos or tissues for in situ hybridization and immunohistochemistry, immunofluorescence microscopy, confocal microscopy, molecular biology, embryological techniques (DNA and RNA injections into zebrafish embryos).The results of the practical course are presented by each student in the form of a written report as well as a talk at the end of the course. The students also take part in the weekly lab meetings where they learn about the ongoing research of all the members of the group. In a Journal Club each student presents a recent publication in the field of their own project.
Educational objectives / Communication
Students learn the basic techniques to study cellular and molecular aspects of developmental genetics (as detailed above). By the end of the course they have been in direct contact with zebrafish or mice and learn how to handle a zebrafish or mouse colony. The students are in an international environment and learn how to write and communicate their results in English.
Dr. Boris Strilic
Contents
This training aims at teaching theoretical knowledge and practical experience in the fields of cellular and molecular biology, more specifically in the fields of endothelial and tumour cell biology. The student(s) will participate in ongoing projects in the lab, including the possibility to work on mice as model organism (depending on the project and availability- under supervision). The student(s) will also participate in regular meetings, and the obtained data will be summarized in a written protocol. The lab-atmosphere is international.Educational objectives / Competences
This training aims at learning different techniques from the above-mentioned fields, including cell culture of cell lines and primary cells, siRNA-mediated knock-down of genes, preparation of histological sections including staining, confocal microscopy and image analysis, PCR, Western blots, immunoprecipitation, etc.
Prof. Dr. Virginie Lecaudey
Contents
The practical course offers basic theoretical and experimental knowledge in the area of developmental cell biology. Principal areas of research are the mechanisms underlying cell migration and morphogenesis during organ formation using the zebrafish. Our main model system is the lateral line, a sensory system present in fish that derives from a group of cells that migrate in a collective manner. The students take part in ongoing experiments in the laboratory to elucidate, for example, the mechanisms underlying cell migration, cell differentiation, cell shape changes or cell proliferation in this system. The techniques used include basic genetics techniques, molecular biology, in situ hybridization and immunohistochemistry as well as handling zebrafish (crossing, injection, genotyping) and confocal and live imaging.The results of the practical course are presented by every student on the form of a written protocol and a talk at the end of the course. The students also take part on the weekly lab meetings where they learn about the ongoing research of the members of the group. In a Journal Club every student presents a recent publication on the field of their own projects.
Educational objectives / competences
Students learn the basic techniques of molecular and developmental biology including zebrafish handling and modern live imaging techniques as detailed above. By the end of the course they have been in direct contact with mice and learn how to handle a mouse colony. The students are in an international environment and learn how to write and communicate their results in English.
Prof. Dr. Michaela Mueller-McNicoll
Contents
In the practical course basics in RNA Biology and animal cell culture will be learned. The students will learn and perform the analysis of RNA expression and RNA-protein-interactions in cells. The students will work under supervision on their own scientific project which is leaned on the scientific work and ongoing experiments of the working group of AK Müller-McNicoll. The experimental results will be presented in a seminar and in form of a graded protocol. During the practical course the students learn to work with different eukaryotic cell lines, the production and transfection of siRNAs to knockdown specific proteins of interest, the purification of RNA-binding proteins from cells and the identification and quantification of bound RNAs via quantitative RT-PCR or other methods.Other techniques comprise immunofluorescence microscopy, PCR, western Blotting, DNA-cloning, mutagenesis of proteins, subcellular fractionation and differentiating of cells.
Educational objectives / Competences
Students will be familiarized with scientific literature; will have additional knowledge in RNA biology and special methods of transcript analysis.
They will get practical experience in sterile working with cells and their analysis. At the end of the course they will be able to work with cell cultures for further analysis. They learn how to write, communicate and present their results in English language.
Prof. Dr. Manfred Koessl & Dr. Julio Hechavarria
Contents
The main goal of this course is to understand how mammals communicate using acoustic information (sounds). The course is designed from the perspective of the “broadcaster-receiver" approach, and therefore it is consequently subdivided into two parts. The first part is meant for understanding the sounds broadcasted by two mammalian species (Mongolian gerbils and bats) while they are communicating. Basically, using bioacoustics tools, the students will try to figure out the vocal alphabet of bats and gerbils. The second part of the course deals with the receiver. In this part, the students will learn how the gerbil's voice is processed in the brain by neurons located in the auditory cortex. The main aim here is to assess what happens in the brain when an animal hears a behaviourally relevant sound. At the beginning of each course part, there will be introductory discussions that will provide the students with the necessary theoretical background for conducting and understanding the different experiments. An introduction to statistics and to MATLAB will also be offered. The final report will be written in the form of a scientific paper, and the results will be presented in the form of a short talk.Educational objectives / Competences
By the end of the course, the students should be able to: (1) Understand basic concepts of bioacoustics such as the sound as a mechanical wave, sound transduction using microphones, analogue-to-digital conversion using sound cards. (2) Measure basic parameters of a sound wave (frequency, duration, intensity). (3) Perform basic surgeries required for acquiring neuronal data. (4) Understand basic neuroscience concepts such as: action potential, local field potential, receptive field, brain topography,
spike clustering, brain oscillations. (5) Testing hypothesis using basic statistical tests (normality tests, parametric and non-parametric t-tests and analyses of variance (ANOVA)).
Prof. Dr. Amparo Acker-Palmer & Dr. Bettina Kirchmaier
Contents
The practical course offers basic theoretical and experimental knowledge in the area of cellular, molecular and systemic neurobiology in mouse and zebrafish. The students work on their own projects under supervision and present the results in the form of a seminar talk. In a second seminar talk they present an original publication from the field of their projects. By writing a result protocol, they will learn how to write scientific reports.The practical course is divided in two units. The first part includes the following tasks: basic mouse genetic techniques, processing of brain tissue for immunohistochemistry, basic techniques of working with neuronal cell cultures, immunofluorescence microscopy, confocal microscopy, and biochemical techniques including protein gel electrophoresis and Western blotting. In the second part of the practical course, the students will be introduced to basic zebrafish genetics using methods in molecular biology and histological techniques, confocal microscopy and brightfield microscopy as well as zebrafish embryo manipulation and basic behavioural tests.
Educational objectives / Competences
Students learn the basic techniques for studying cellular, molecular, and systemic neurobiology (as detailed above). They work with cultured cells under sterile conditions, with the epifluorescence microscope and the stereo microscope. The students will be trained in zebrafish embryo handling and basic genetic techniques, and quantify and analyse the obtained data and images. The students are in an international environment and learn how to write and communicate their results in English.
Prof. Dr. Franziska Matthaeus
Contents
This module provides a foundation of theoretical biology in the form of individual student research projects, mainly based on examples of cell migration. Methods and techniques, which are hereby acquired and applied, range from data or image analysis (such as segmentation, tracking, dimension reduction of large data sets, or network analysis), to the modelling and simulation of motile cells. Examples of projects involving modelling and simulation are collective migration (of cancer cells or in developmental processes), chemotaxis, pattern formation, or the description of chemical or mechanical regulatory mechanisms of motility. Alternatively, data analysis, modelling or simulation can be carried out based on a project providing suitable data collected during a previous module.Image analysis will mostly be carried out using conventional software tools (ImageJ, Matlab). For post-processing of the data, visualization or statistical analysis the student will be supported in the generation of own software. The students may code in the programming language of their choice. For beginners, an introduction to programming (in Python, Matlab or Julia) is provided. Also modelling and simulation will involve the generation of code. The topic and results of the research project will be presented in the group seminar. In addition, every student will submit a protocol comprising 10 – 30 pages.
Educational objectives / Competences
The goal of the module is to provide and extend programming skills, data analysis methods and modelling approaches for dynamical or spatio-temporal processes, or large data sets. Students should be able to apply these methods/approaches in their future research and use them as a foundation for further development. Independent research, usage of original literature, and scientific writing will be strengthened.
Prof. Dr. Stefan Eimer
Contents
In this practical course, students learn how to use the multicellular model organism C. elegans to investigate the early molecular mechanisms and cellular alterations leading to the development of Parkinson's disease. The inactivation and analysis of homologous genes that are associated with the hereditary form of Parkinson's disease, attempts to understand the function of these genes in normal cells. In addition, it will be investigated, which cellular alterations result from the inactivation of these genes and what conclusions can be drawn on possible early mechanisms of Parkinson's disease development.The generated mutant animals are genetically, biochemically and cell biologically examined using high-resolution microscopy techniques such as confocal microscopy and electron microscopy. The students learn how to use the model system C. elegans and the latest methods of genetic manipulation such as CRISPR/Cas9-mediated genome editing or RNAi gene knockdown as well as quantitative evaluation of microscopy data. The students present their results in a seminar talk and in the form of a graded protocol.
Educational objectives / Competences
During the internship, students learn how genetic manipulation and analysis of a multicellular model organism can help to understand complex cellular relationships that can lead to the development of neurodegenerative disorders.
Prof. Dr. Jasmin Hefendehl
Contents
The practical course offers basic theoretical and experimental knowledge in the area of neurodegenerative and vascular disorders. The students work on their own projects under supervision and present the results in the form of a seminar talk. This talk also includes an original publication from the field of their projects. By writing a result protocol, they will learn how to write scientific reports.The practical course consists of systemic, cellular and molecular aspects that will be addressed. This includes the following techniques: in vivo 2-Photon Microscopy, Image- and data analysis, basic mouse genetic techniques, processing of brain tissue for immunohistochemistry, basic primary cell culture techniques, immunofluorescence microscopy, confocal microscopy as well as biochemical techniques including protein gel electrophoresis and Western blotting.
Educational objectives / Competences
In this elective module, the student will learn fundamental techniques used to in the research area of neurodegenerative disorders (as described above) by using the mouse model organism. In vivo 2-Photon imaging enables us to record systemic as well as cellular processes in real time. The students are presented with the opportunity to observe in vivo animal handling and the live imaging process. The acquired data will be analyzed by the students, teaching them basic Image- and data analysis skills. The immunohistochemical stainings of brain sections will teach the students the technique as well as the underlying scientific question of the experiment. Moreover, the students will work with cultured cells under sterile conditions, with the epifluorescence – and stereo microscope. The students are in an international environment and learn how to write and communicate their results in English.
Dr. Florian Freudenberg
Contents
In this practical course the molecular and cellular causes of psychiatric disorders will be explored. Students will be introduced to a broad spectrum of translational methods. These include cell culture techniques for the functional study of candidate genes (including the production of primary cell cultures from humans and/or mice, production of viral vectors and viral gene transfer), molecular and cell biological mechanisms in the cell model and/or optionally in the mouse model (including behavioural biology in mice that have been genetically modified and/or pharmacologically treated). Following such experiments, various molecular biological (including quantitative PCR, Western blot, ELISA), neuroanatomical (brain preparation, cryostat cutting, staining methods), immunohistochemical and microscopic (including fluorescence microscopy with and without structured illumination) characterisations will be performed. It is also possible to gain insight into behavioral studies and imaging techniques (functional magnetic resonance imaging [fMRT], electroencephalography [EEG], magnetoencephalography [MEG]) used to evaluate abnormal neuronal processing in human psychiatric disorders.The experiments are carried out in the laboratories of the Department of Psychiatry, Psychosomatics and Psychotherapy of the University Hospital Frankfurt. Current projects of the department will be carried out under supervision. The results will be presented in a presentation in the lab seminar and documented in the form of a written report.
Educational objectives / Competences
Students can plan, carry out and analyse experiments used to investigate psychiatric disorders. Students learn and develop scientific approaches and literature research. The students document their results and communicate them in oral and written form. In a series of seminars (including the opportunity to participate in case presentations), students also acquire basic knowledge of the psychiatric disorders studied and are able to describe them.
Dr. Phillip Grote
Contents
This course serves as an introduction into the basic theoretical and experimental field of cardiovascular development and their implications in congenital heart disease. The main model that is used in our lab is the mouse model system, which is complemented with cell biology work in primary cardiac cells isolated from mice or work with cell lines in vitro. The student takes over a subproject of the ongoing research in the lab. The focus of the lab is the contribution of long non-protein coding RNAs (lncRNAs) to various cell regulatory process in the cardiac system. To this end, we genetically engineer mouse embryonic stem cells with CRISPR/Cas9 to generate genetic mutants of lncRNAs in heart development. In addition, we use other CRISPR technologies (e.g. CRISPRa (gene activation), CRISPRi (gene inactivation) and Cas13 (RNA inhibition) to modulate gene programs in either ex vivo or in vitro settings. Such engineered stem cells are differentiated into early cardiac cells in vitro or directly used to generate mouse embryos by our own pipeline. Any impact on cardiac development will be observed.At the end of the lab rotation the students will prepare a short, written report and will present their finding in the general lab meeting to the fellow lab members for discussion and possible future research direction. Students will participate in the weekly lab meetings and will get familiar with the ongoing research in the lab.
Educational objectives / Competences
The students will learn how to use various CRISPR systems to dissect gene regulatory programs in the cardiovascular system and related cells, which of course can be applied to any other field of research. They learn how to ‘read’ and interpret whole genome data to direct hypothesis on gene regulation and how to design genetic manipulation experiments to verify them.
Dr. Stefan Momma
Contents
Extracellular vesicles are released membrane vesicles from all cells, which contain functional proteins as well as nucleic acids. The communication between cells via extracellular vesicles is a relative new and complex field of research.In the practical course, the students will get an introduction to extracellular vesicles.
Main points are the aspects of purification, classification, analysis of RNA-Protein content as well as visualization and transfer analysis of functional molecules between cell populations in vitro and in vivo.
The students will work with different cell culture techniques, immunofluorescence microscopy, flow cytometric analysis and other related techniques.
Educational objectives / Competences
The students will get knowledge and first experience in the field of Biology with extracellular vesicles and the communication via extracellular vesicles. They learn basic techniques to work with vesicles and they will learn to analyze the biological function. Students will be familiarized with scientific literature and learn how to write, communicate and present their results in English.
Prof. Dr. Andreas Geburtig-Chiocchetti
Contents
In this laboratory practical course, the fundamentals are taught to decipher the molecular genetic mechanisms behind neuropsychiatric disorders such as autism or social behavior disorder. On the one hand, bioinformatic methods can be learned to identify genetic markers for the disorders or to link transcriptome data from neuronal cell models to patient data in such a way that new insights into the cause of the disorder or biologically defined subgroups of patients can be identified. In particular, omics analyses as well as machine learning methods are used here. On the other hand, neuronal human cell models are generated based on the identified genetic markers (CRISPTR/Cas9). Here, the students learn how to handle human cell models, especially human embryonic stem cells (hESC), induced pluripotent stem cells and especially the cultivation and quantitative evaluation (real-time PCR, confocal microscopy) of neuronal progenitor cells, differentiated neurons and cerebral organoids. The participant will discuss the results in our scientific seminars.Educational objectives / Competences
The practical course teaches students how to implement translational research approaches. In particular, they learn how to use molecular genetic and bioinformatics methods to link findings from the cell model with patient data and vice versa.
- Studying at Goethe University
- International applicants
- Faculties
- Overview of study programmes
- Programme for refugees
- GRADE
- Goethe Business School (continuing education)
- Research at Goethe University
- Scientific news
- Goethe Welcome Center (for international researchers)
- Collaborative research projects
- Individual research
- Visiting fellowships
- Endowed chairs
- About the University
- News-in-brief
- University administration
- Campus locations
- Campus life
- University archives (German)
- Rhine-Main-Universities





















